matchms.Spectrum module¶
- class matchms.Spectrum.Spectrum(mz: array, intensities: array, metadata: dict | None = None, metadata_harmonization: bool = True)[source]¶
Bases:
objectContainer for a collection of peaks, losses and metadata.
Spectrum peaks are stored as
Fragmentsobject which can be addressed calling spectrum.peaks and contains m/z values and the respective peak intensities.Spectrum metadata is stored as
Metadataobject which can be addressed by spectrum.metadata.Code example
import numpy as np from matchms import Scores, Spectrum from matchms.similarity import CosineGreedy spectrum = Spectrum(mz=np.array([100, 150, 200.]), intensities=np.array([0.7, 0.2, 0.1]), metadata={"id": 'spectrum1', "precursor_mz": 222.333, "peak_comments": {200.: "the peak at 200 m/z"}}) print(spectrum) print(spectrum.peaks.mz[0]) print(spectrum.peaks.intensities[0]) print(spectrum.get('id')) print(spectrum.peak_comments.get(200))
Should output
Spectrum(precursor m/z=222.33, 3 fragments between 100.0 and 200.0) 100.0 0.7 spectrum1 the peak at 200 m/z
- losses¶
Losses of spectrum, the difference between the precursor and all peaks.
Can be filled with
from matchms import Fragments spectrum.losess = Fragments(mz=np.array([50.]), intensities=np.array([0.1]))
- Type:
Fragments or None
- __init__(mz: array, intensities: array, metadata: dict | None = None, metadata_harmonization: bool = True)[source]¶
- Parameters:
mz – Array of m/z for the peaks
intensities – Array of intensities for the peaks
metadata – Dictionary with for example the scan number of precursor m/z.
metadata_harmonization (bool, optional) – Set to False if default metadata filters should not be applied. The default is True.
- get(key: str, default=None)[source]¶
Retrieve value from
metadatadict. Shorthand forval = self.metadata[key]
- metadata_dict(export_style: str = 'matchms') dict[source]¶
Convert spectrum metadata to Python dictionary.
- Parameters:
export_style – Converts the keys to the required export style. One of [“matchms”, “massbank”, “nist”, “riken”, “gnps”]. Default is “matchms”
- metadata_hash()[source]¶
Return a (truncated) sha256-based hash which is generated based on the spectrum metadata. Spectra with same metadata results in same metadata_hash.
- plot(figsize=(8, 6), dpi=200, **kwargs)[source]¶
Plot to visually inspect a spectrum run
spectrum.plot()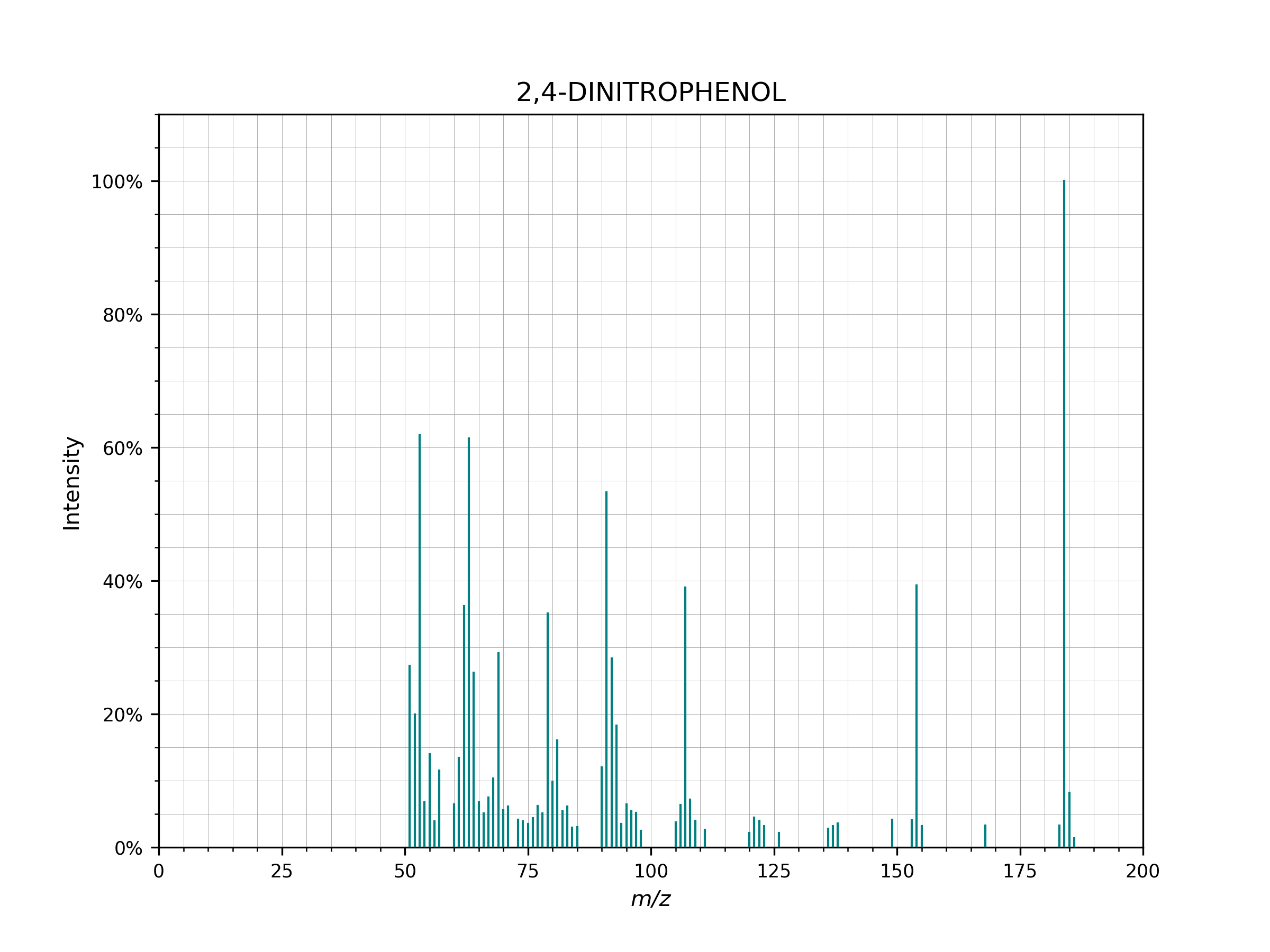
Example of a spectrum plotted using
spectrum.plot()..¶
- plot_against(other_spectrum, figsize=(8, 6), dpi=200, **spectrum_kws)[source]¶
Compare two spectra visually in a mirror plot.
To visually compare the peaks of two spectra run
spectrum.plot_against(other_spectrum)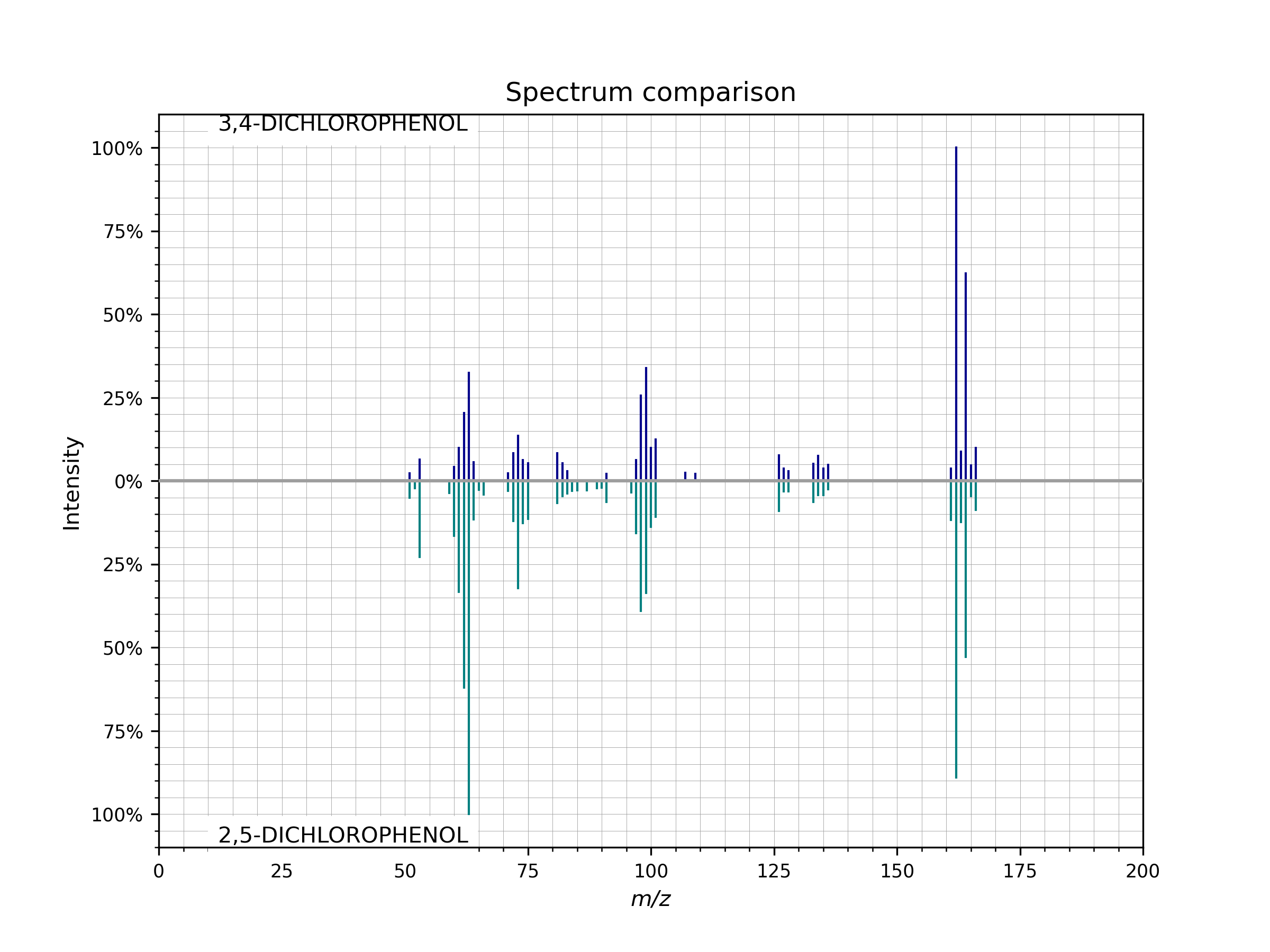
Example of a mirror plot comparing two spectra
spectrum.plot_against()..¶
- spectrum_hash()[source]¶
Return a (truncated) sha256-based hash which is generated based on the spectrum peaks (mz:intensity pairs). Spectra with same peaks will results in same spectrum_hash.
