matchms.filtering package¶
Processing (or: filtering) mass spectra¶
Provided functions will usually only perform a single action to a spectrum. This can be changes or corrections of metadata, or peak filtering. More complicated processing pipelines can be build by stacking several of the provided filters.
Because there are numerous filter functions in matchms and because they often need to be applied in a specific order, the most feasible workflow for users is to use the SpectrumProcessor class to define a spetrum processing pipeline. Here is an example:
import numpy as np
from matchms import Spectrum
from matchms import SpectrumProcessor
spectrum = Spectrum(mz=np.array([100, 120, 150, 200.]),
intensities=np.array([200.0, 300.0, 50.0, 1.0]),
metadata={'id': 'spectrum1'})
# Users can pick a predefined pipeline ("basic", "default", "fully_annotated")
# Or set to None if no predefined settings are desired.
processing = SpectrumProcessor("basic")
# Additional filters can be added as desired
processing.add_matchms_filter("normalize_intensities")
# Run the processing pipeline:
spectrum_filtered = processing.process_spectrum(spectrum)
max_intensity = spectrum_filtered.peaks.intensities.max()
print(f"Maximum intensity is {max_intensity:.2f}")
Should output
Maximum intensity is 1.00
It is also possible to run each filter function individually. This for instance makes sense if users want to develop a highly customized spectrum processing routine. Example of how to use a single filter function:
import numpy as np
from matchms import Spectrum
from matchms.filtering import normalize_intensities
spectrum = Spectrum(mz=np.array([100, 120, 150, 200.]),
intensities=np.array([200.0, 300.0, 50.0, 1.0]),
metadata={'id': 'spectrum1'})
spectrum_filtered = normalize_intensities(spectrum)
max_intensity = spectrum_filtered.peaks.intensities.max()
print(f"Maximum intensity is {max_intensity:.2f}")
Should output
Maximum intensity is 1.00
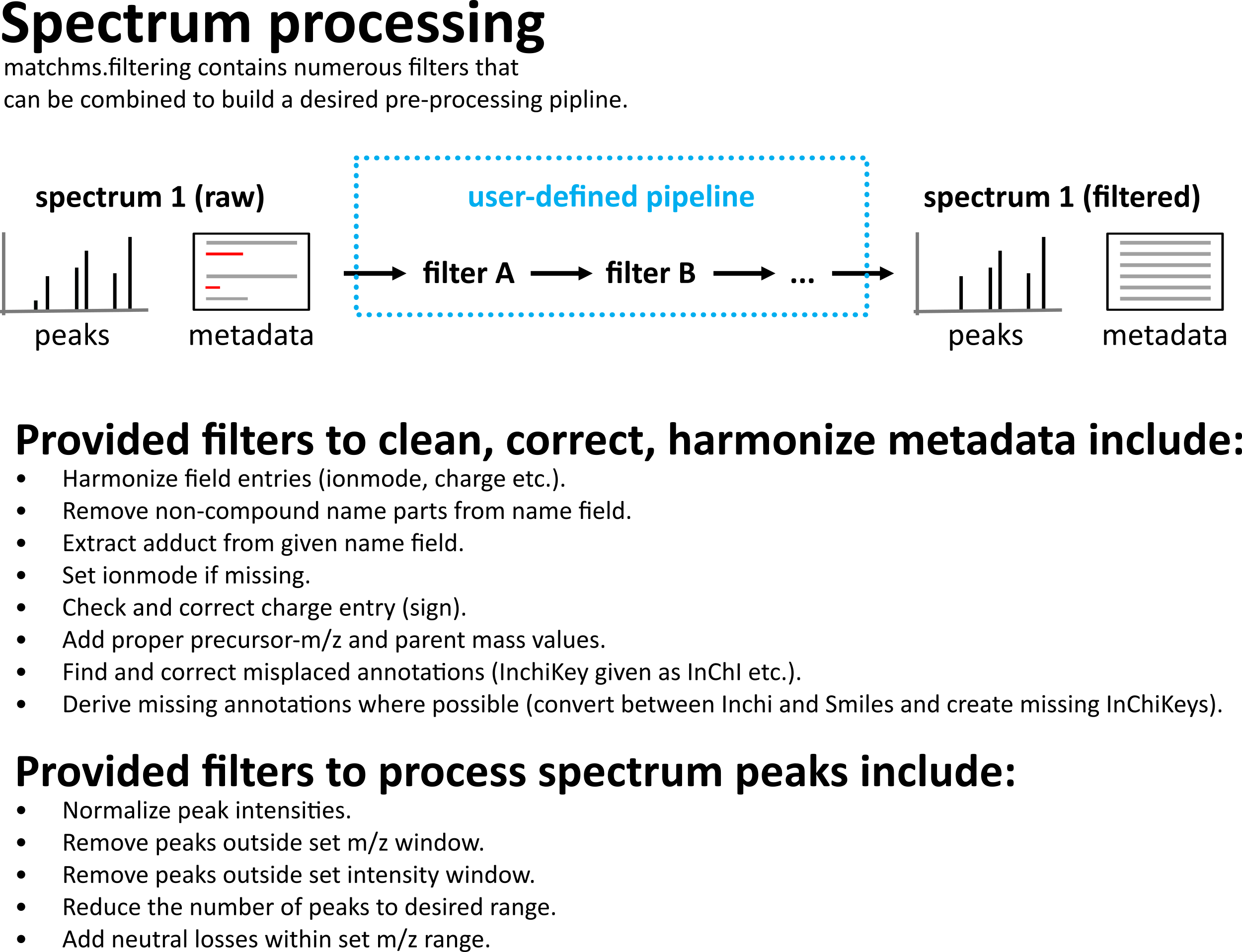
Sketch of matchms spectrum processing.¶
- class matchms.filtering.SpeciesString(dirty: str)[source]¶
Bases:
objectA class to process and clean different types of chemical structure strings including InChI, InChIKey, and SMILES.
The class takes a raw input string, determines the intended structure type, and then cleans the string based on its type.
- target¶
The intended structure type determined from the input string. Could be ‘inchi’, ‘inchikey’, ‘smiles’, or None if no valid type was identified.
- Type:
- matchms.filtering.add_compound_name(spectrum_in: Spectrum) Spectrum[source]¶
Add compound_name to correct field: “compound_name” in metadata.
- matchms.filtering.add_fingerprint(spectrum_in: Spectrum | None, fingerprint_type: str = 'daylight', nbits: int = 2048) Spectrum | None[source]¶
Add molecular finterprint to spectrum.
If smiles or inchi present in metadata, derive a molecular finterprint and add it to the spectrum.
- Parameters:
spectrum_in – Input spectrum.
fingerprint_type – Determine method for deriving molecular fingerprints. Supported choices are “daylight”, “morgan1”, “morgan2”, “morgan3”. Default is “daylight”.
nbits – Dimension or number of bits of generated fingerprint. Default is 2048.
- matchms.filtering.add_losses(spectrum_in: Spectrum, loss_mz_from=0.0, loss_mz_to=1000.0) Spectrum[source]¶
Derive losses based on precursor mass.
- Parameters:
spectrum_in – Input spectrum.
loss_mz_from – Minimum allowed m/z value for losses. Default is 0.0.
loss_mz_to – Maximum allowed m/z value for losses. Default is 1000.0.
- matchms.filtering.add_parent_mass(spectrum_in: Spectrum, estimate_from_adduct: bool = True, overwrite_existing_entry: bool = False) Spectrum[source]¶
Add estimated parent mass to metadata (if not present yet).
Method to calculate the parent mass from given precursor m/z together with charge and/or adduct. Will take precursor m/z from “precursor_mz” as provided by running add_precursor_mz. For estimate_from_adduct=True this function will estimate the parent mass based on the mass and charge of known adducts. The table of known adduct properties can be found under
matchms/data/known_adducts_table.csv.- Parameters:
spectrum_in – Input spectrum.
estimate_from_adduct – When set to True, use adduct to estimate actual molecular mass (“parent mass”). Default is True. Switches back to charge-based estimate if adduct does not match a known adduct.
overwrite_existing_entry – Default is False. If set to True, a newly computed value will replace existing ones.
- matchms.filtering.add_precursor_mz(spectrum_in)[source]¶
Add precursor_mz to correct field and make it a float.
For missing precursor_mz field: check if there is “pepmass”” entry instead. For string parsed as precursor_mz: convert to float.
- matchms.filtering.add_retention_index(spectrum_in: Spectrum) Spectrum[source]¶
Add retention index into ‘retention_index’ key if present.
- Parameters:
spectrum – Spectrum with RI information.
- Return type:
Spectrum with RI info stored under ‘retention_index’.
- matchms.filtering.add_retention_time(spectrum_in: Spectrum) Spectrum[source]¶
Add retention time information to the ‘retention_time’ key as float. Negative values and those not convertible to a float result in ‘retention_time’ being ‘None’.
- Parameters:
spectrum – Spectrum with retention time information.
- Return type:
Spectrum with harmonized retention time information.
- matchms.filtering.clean_adduct(spectrum_in)[source]¶
Clean adduct and make it consistent in style. Will transform adduct strings of type ‘M+H+’ to ‘[M+H]+’.
- Parameters:
spectrum_in – Matchms Spectrum object.
- matchms.filtering.clean_compound_name(spectrum_in: Spectrum) Spectrum[source]¶
Clean compound name.
A list of frequently seen name additions that do not belong to the compound name will be removed.
- matchms.filtering.correct_charge(spectrum_in: Spectrum) Spectrum[source]¶
Correct charge values based on given ionmode.
For some spectrums, the charge value is either undefined or inconsistent with its ionmode, which is corrected by this filter.
- Parameters:
spectrum_in – Input spectrum.
- matchms.filtering.default_filters(spectrum: Spectrum) Spectrum[source]¶
Collection of filters that are considered default and that do no require any (factory) arguments.
Collection is
- matchms.filtering.derive_adduct_from_name(spectrum_in: Spectrum, remove_adduct_from_name: bool = True) Spectrum | None[source]¶
Find adduct in compound name and add to metadata (if not present yet).
Method to interpret the given compound name to find the adduct.
- Parameters:
spectrum_in – Input spectrum.
remove_adduct_from_name – Remove found adducts from compound name if set to True. Default is True.
- matchms.filtering.derive_annotation_from_compound_name(spectrum_in: <module 'matchms.Spectrum' from '/home/docs/checkouts/readthedocs.org/user_builds/matchms/checkouts/latest/readthedocs/../matchms/Spectrum.py'>, annotated_compound_names_file: str | None = None, mass_tolerance: float = 0.1)[source]¶
Adds smiles, inchi, inchikey based on compound name by searching pubchem
This filter is only run, if there is not yet a valid smiles or inchi in the metadata. The smiles, inchi and inchikey are only added if the found annotation is close enough to the parent mass.
- Parameters:
spectrum_in – The input spectrum.
annotated_compound_names_file (Optional[str]) – Any compound name that was searched for on pubchem will be added to this file. If a compound name is already in this file it will be used instead of looking up at pubchem. This file can be reused for future runs, speeding up the process. If None. The compound names found will still be cached for this run, but won’t be reusable for future runs. The csv file should contain the columns [“compound_name”, “smiles”, “inchi”, “inchikey”, “monoisotopic_mass”]
mass_tolerance – Acceptable mass difference between query compound and pubchem result.
- matchms.filtering.derive_formula_from_name(spectrum_in: Spectrum, remove_formula_from_name: bool = True) Spectrum[source]¶
Detect and remove misplaced formula in compound name and add to metadata.
Method to find misplaced formulas in compound name based on regular expression. This will not chemically test the detected formula, so the search is limited to frequently occuring types of shape ‘C47H83N1O8P1’.
- Parameters:
spectrum_in – Input spectrum.
remove_formula_from_name – Remove found formula from compound name if set to True. Default is True.
- matchms.filtering.derive_inchi_from_smiles(spectrum_in: Spectrum) Spectrum[source]¶
Find missing Inchi and derive from smiles where possible.
- matchms.filtering.derive_inchikey_from_inchi(spectrum_in: Spectrum) Spectrum[source]¶
Find missing InchiKey and derive from Inchi where possible.
- matchms.filtering.derive_ionmode(spectrum_in: Spectrum) Spectrum[source]¶
Derive missing ionmode based on adduct.
Some input formates (e.g. MGF files) do not always provide a correct ionmode. This function reads the adduct from the metadata and uses this to fill in the correct ionmode where missing.
- Parameters:
spectrum – Input spectrum.
- Return type:
Spectrum object with ionmode attribute set.
- matchms.filtering.derive_smiles_from_inchi(spectrum_in: Spectrum) Spectrum[source]¶
Find missing smiles and derive from Inchi where possible.
- matchms.filtering.harmonize_undefined_inchi(spectrum_in: Spectrum, undefined: str = '', aliases: List[str] | None = None) Spectrum[source]¶
Replace all aliases for empty/undefined inchi entries by value of
undefinedargument.- Parameters:
undefined – Give desired entry for undefined inchi fields. Default is “”.
aliases – Enter list of strings that are expected to represent undefined entries. Default is [“”, “N/A”, “NA”, “n/a”].
- matchms.filtering.harmonize_undefined_inchikey(spectrum_in: Spectrum, undefined: str = '', aliases: List[str] | None = None) Spectrum[source]¶
Replace all aliases for empty/undefined inchikey entries by
undefined.- Parameters:
undefined – Give desired entry for undefined inchikey fields. Default is “”.
aliases – Enter list of strings that are expected to represent undefined entries. Default is [“”, “N/A”, “NA”, “n/a”, “no data”].
- matchms.filtering.harmonize_undefined_smiles(spectrum_in: Spectrum, undefined: str = '', aliases: List[str] | None = None) Spectrum[source]¶
Replace all aliases for empty/undefined smiles entries by
undefined.- Parameters:
undefined – Give desired entry for undefined smiles fields. Default is “”.
aliases – Enter list of strings that are expected to represent undefined entries. Default is [“”, “N/A”, “NA”, “n/a”, “no data”].
- matchms.filtering.interpret_pepmass(spectrum_in)[source]¶
Reads pepmass field (if present) and adds values to correct field(s).
The field “pepmass” or “PEPMASS” is often used to describe the precursor ion. This function will interpret the values as (mz, intensity, charge) tuple. Those will be splitted (if present) added to the fields “precursor_mz”, “precursor_intensity”, and “charge”.
- matchms.filtering.make_charge_int(spectrum_in: Spectrum) Spectrum[source]¶
Convert charge field to integer (if possible).
- matchms.filtering.normalize_intensities(spectrum_in: Spectrum) Spectrum[source]¶
Normalize intensities of peaks (and losses) to unit height.
- matchms.filtering.reduce_to_number_of_peaks(spectrum_in: Spectrum, n_required: int = 0, n_max: int = inf, ratio_desired: float | None = None) Spectrum[source]¶
Lowest intensity peaks will be removed when it has more peaks than desired.
- Parameters:
spectrum_in – Input spectrum.
n_required – Number of minimum required peaks. Spectra with fewer peaks will be set to ‘None’. Default is 1.
n_max – Maximum number of peaks. Remove peaks if more peaks are found. Default is inf.
ratio_desired – Set desired ratio between maximum number of peaks and parent mass. For spectra without parent mass (e.g. GCMS spectra) this will raise an error when ratio_desired is used. Default is None.
- matchms.filtering.remove_peaks_around_precursor_mz(spectrum_in: Spectrum, mz_tolerance: float = 17) Spectrum[source]¶
- Remove peaks that are within mz_tolerance (in Da) of
the precursor mz, exlcuding the precursor peak.
- Parameters:
spectrum_in – Input spectrum.
mz_tolerance – Tolerance of mz values that are not allowed to lie within the precursor mz. Default is 17 Da.
- matchms.filtering.remove_peaks_outside_top_k(spectrum_in: Spectrum, k: int = 6, mz_window: float = 50) Spectrum[source]¶
- Remove all peaks which are not within mz_window of at least one
of the k highest intensity peaks of the spectrum.
- Parameters:
spectrum_in – Input spectrum.
k – The number of most intense peaks to compare to. Default is 6.
mz_window – Window of mz values (in Da) that are allowed to lie within the top k peaks. Default is 50 Da.
- matchms.filtering.repair_adduct_based_on_smiles(spectrum_in: <module 'matchms.Spectrum' from '/home/docs/checkouts/readthedocs.org/user_builds/matchms/checkouts/latest/readthedocs/../matchms/Spectrum.py'>, mass_tolerance: float, accept_parent_mass_is_mol_wt: bool = True)[source]¶
Corrects the adduct of a spectrum based on its SMILES representation and the precursor m/z.
Given a spectrum, this function tries to match the spectrum’s parent mass, derived from its precursor m/z and known adducts, to the neutral monoisotopic mass of the molecule derived from its SMILES representation. If a match is found within a given mass tolerance, the adduct and parent mass of the spectrum are updated.
Parameters:¶
- spectrum_inSpectrum
The input spectrum whose adduct needs to be repaired.
- mass_tolerancefloat
Maximum allowed mass difference between the calculated parent mass and the neutral monoisotopic mass derived from the SMILES.
- accept_parent_mass_is_mol_wtbool, optional (default=True)
Allows the function to attempt repairing the spectrum’s parent mass by assuming it represents the molecule’s weight. If True, further checks and corrections are made using repair_parent_mass_is_mol_wt.
- matchms.filtering.repair_inchi_inchikey_smiles(spectrum_in: Spectrum) Spectrum[source]¶
Check if inchi, inchikey, and smiles entries seem correct. Detect and correct if any of those entries clearly belongs into one of the other two fields (e.g. inchikey found in inchi field).
- matchms.filtering.repair_not_matching_annotation(spectrum_in: <module 'matchms.Spectrum' from '/home/docs/checkouts/readthedocs.org/user_builds/matchms/checkouts/latest/readthedocs/../matchms/Spectrum.py'>)[source]¶
Repairs mismatches in a spectrum’s annotations related to SMILES, InChI, and InChIKey.
Given a spectrum, this function ensures that the provided SMILES, InChI, and InChIKey annotations are consistent with one another. If there are discrepancies, they are resolved as follows:
- If the SMILES and InChI do not match:
Both SMILES and InChI are checked against the parent mass.
The annotation that matches the parent mass is retained, and the other is regenerated.
- If the InChIKey does not match the InChI:
A new InChIKey is generated from the InChI and replaces the old one.
Warnings and information logs are generated to track changes and potential issues. For correctness of InChIKey entries, only the first 14 characters are considered.
Parameters:¶
- spectrum_inSpectrum
The input spectrum containing annotations to be checked and repaired.
Returns:¶
- Spectrum
A cloned version of the input spectrum with corrected annotations. If the input spectrum is None, it returns None.
- matchms.filtering.repair_parent_mass_is_mol_wt(spectrum_in: <module 'matchms.Spectrum' from '/home/docs/checkouts/readthedocs.org/user_builds/matchms/checkouts/latest/readthedocs/../matchms/Spectrum.py'>, mass_tolerance: float)[source]¶
Changes the parent mass from molecular mass into monoistopic mass
Manual entered precursor mz is sometimes wrongly added as Molar weight instead of monoisotopic mass
- matchms.filtering.repair_parent_mass_match_smiles_wrapper(spectrum_in: Spectrum, mass_tolerance: float = 0.2) Spectrum | None[source]¶
Wrapper function for repairing a mismatch between parent mass and smiles mass
- matchms.filtering.repair_precursor_is_parent_mass(spectrum_in: <module 'matchms.Spectrum' from '/home/docs/checkouts/readthedocs.org/user_builds/matchms/checkouts/latest/readthedocs/../matchms/Spectrum.py'>, mass_tolerance)[source]¶
Repairs parent mass and precursor mz if the parent mass is entered instead of the precursor_mz
- matchms.filtering.repair_smiles_of_salts(spectrum_in, mass_tolerance)[source]¶
Repairs the smiles of a salt to match the parent mass. E.g. C1=NC2=NC=NC(=C2N1)N.Cl is converted to 1=NC2=NC=NC(=C2N1)N if this matches the parent mass Checks if parent mass matches one of the ions
- matchms.filtering.require_correct_ionmode(spectrum_in: Spectrum, ion_mode_to_keep) Spectrum | None[source]¶
Validates the ion mode of a given spectrum. If the spectrum’s ion mode doesn’t match the ion_mode_to_keep, it will be removed and a log message will be generated.
- Parameters:
- Returns:
The validated spectrum if its ion mode matches the desired one, or None otherwise.
- Return type:
Spectrum or None
- matchms.filtering.require_minimum_number_of_high_peaks(spectrum_in: Spectrum, no_peaks: int = 5, intensity_percent: float = 2.0) Spectrum[source]¶
- Returns None if the number of peaks with relative intensity
above or equal to intensity_percent is less than no_peaks.
- Parameters:
spectrum_in – Input spectrum.
no_peaks – Minimum number of peaks allowed to have relative intensity above intensity_percent. Less peaks will return none. Default is 5.
intensity_percent – Minimum relative intensity (as a percentage between 0-100) for peaks that are searched. Default is 2
- matchms.filtering.require_minimum_number_of_peaks(spectrum_in: Spectrum, n_required: int = 10, ratio_required: float | None = None) Spectrum[source]¶
Spectrum will be set to None when it has fewer peaks than required.
- Parameters:
spectrum_in – Input spectrum.
n_required – Number of minimum required peaks. Spectra with fewer peaks will be set to ‘None’.
ratio_required – Set desired ratio between minimum number of peaks and parent mass. Default is None.
- matchms.filtering.require_parent_mass_match_smiles(spectrum_in: Spectrum, mass_tolerance) Spectrum | None[source]¶
Validates if the parent mass of the given spectrum matches the mass calculated from its associated SMILES string within a specified tolerance.
- Parameters:
- Returns:
The validated spectrum if its parent mass matches the SMILES mass within the specified tolerance, or None otherwise.
- Return type:
Spectrum or None
- matchms.filtering.require_precursor_below_mz(spectrum_in: Spectrum, max_mz: float = 1000) Spectrum[source]¶
- Returns None if the precursor_mz of a spectrum is above
max_mz.
- Parameters:
spectrum_in – Input spectrum.
max_mz – Maximum mz value for the precursor mz of a spectrum. All precursor mz values greater or equal to this will return none. Default is 1000.
- matchms.filtering.require_precursor_mz(spectrum_in: Spectrum, minimum_accepted_mz: float = 10.0) Spectrum | None[source]¶
Returns None if there is no precursor_mz or if <= minimum_accepted_mz
- Parameters:
spectrum_in – Input spectrum.
minimum_accepted_mz – Set to minimum acceptable value for precursor m/z. Default is set to 10.0.
- matchms.filtering.require_valid_annotation(spectrum: <module 'matchms.Spectrum' from '/home/docs/checkouts/readthedocs.org/user_builds/matchms/checkouts/latest/readthedocs/../matchms/Spectrum.py'>)[source]¶
Removes spectra that are not fully annotated (correct and matching, smiles, inchi and inchikey)
- matchms.filtering.select_by_intensity(spectrum_in: Spectrum, intensity_from: float = 10.0, intensity_to: float = 200.0) Spectrum[source]¶
Keep only peaks within set intensity range (keep if intensity_from >= intensity >= intensity_to). In most cases it is adviced to use
select_by_relative_intensity()function instead.- Parameters:
intensity_from – Set lower threshold for peak intensity. Default is 10.0.
intensity_to – Set upper threshold for peak intensity. Default is 200.0.
- matchms.filtering.select_by_mz(spectrum_in: Spectrum, mz_from: float = 0.0, mz_to: float = 1000.0) Spectrum[source]¶
Keep only peaks between mz_from and mz_to (keep if mz_from >= m/z >= mz_to).
- Parameters:
mz_from – Set lower threshold for m/z peak positions. Default is 0.0.
mz_to – Set upper threshold for m/z peak positions. Default is 1000.0.
- matchms.filtering.select_by_relative_intensity(spectrum_in: Spectrum, intensity_from: float = 0.0, intensity_to: float = 1.0) Spectrum[source]¶
Keep only peaks within set relative intensity range (keep if intensity_from >= intensity >= intensity_to).
- Parameters:
intensity_from – Set lower threshold for relative peak intensity. Default is 0.0.
intensity_to – Set upper threshold for relative peak intensity. Default is 1.0.
Subpackages¶
- matchms.filtering.filter_utils package
- Submodules
- matchms.filtering.filter_utils.derive_precursor_mz_and_parent_mass module
- matchms.filtering.filter_utils.get_neutral_mass_from_smiles module
- matchms.filtering.filter_utils.interpret_unknown_adduct module
- matchms.filtering.filter_utils.load_known_adducts module
- matchms.filtering.filter_utils.smile_inchi_inchikey_conversions module
- Submodules
- matchms.filtering.metadata_processing package
- Submodules
- matchms.filtering.metadata_processing.add_compound_name module
- matchms.filtering.metadata_processing.add_fingerprint module
- matchms.filtering.metadata_processing.add_parent_mass module
- matchms.filtering.metadata_processing.add_precursor_mz module
- matchms.filtering.metadata_processing.add_retention module
- matchms.filtering.metadata_processing.clean_adduct module
- matchms.filtering.metadata_processing.clean_compound_name module
- matchms.filtering.metadata_processing.correct_charge module
- matchms.filtering.metadata_processing.derive_adduct_from_name module
- matchms.filtering.metadata_processing.derive_annotation_from_compound_name module
- matchms.filtering.metadata_processing.derive_formula_from_name module
- matchms.filtering.metadata_processing.derive_inchi_from_smiles module
- matchms.filtering.metadata_processing.derive_inchikey_from_inchi module
- matchms.filtering.metadata_processing.derive_ionmode module
- matchms.filtering.metadata_processing.derive_smiles_from_inchi module
- matchms.filtering.metadata_processing.harmonize_undefined_inchi module
- matchms.filtering.metadata_processing.harmonize_undefined_inchikey module
- matchms.filtering.metadata_processing.harmonize_undefined_smiles module
- matchms.filtering.metadata_processing.interpret_pepmass module
- matchms.filtering.metadata_processing.make_charge_int module
- matchms.filtering.metadata_processing.repair_adduct_based_on_smiles module
- matchms.filtering.metadata_processing.repair_inchi_inchikey_smiles module
- matchms.filtering.metadata_processing.repair_not_matching_annotation module
- matchms.filtering.metadata_processing.repair_parent_mass_is_mol_wt module
- matchms.filtering.metadata_processing.repair_parent_mass_match_smiles_wrapper module
- matchms.filtering.metadata_processing.repair_precursor_is_parent_mass module
- matchms.filtering.metadata_processing.repair_smiles_of_salts module
- matchms.filtering.metadata_processing.require_correct_ionmode module
- matchms.filtering.metadata_processing.require_parent_mass_match_smiles module
- matchms.filtering.metadata_processing.require_precursor_below_mz module
- matchms.filtering.metadata_processing.require_precursor_mz module
- matchms.filtering.metadata_processing.require_valid_annotation module
- Submodules
- matchms.filtering.peak_processing package
- Submodules
- matchms.filtering.peak_processing.add_losses module
- matchms.filtering.peak_processing.normalize_intensities module
- matchms.filtering.peak_processing.reduce_to_number_of_peaks module
- matchms.filtering.peak_processing.remove_peaks_around_precursor_mz module
- matchms.filtering.peak_processing.remove_peaks_outside_top_k module
- matchms.filtering.peak_processing.require_minimum_number_of_high_peaks module
- matchms.filtering.peak_processing.require_minimum_number_of_peaks module
- matchms.filtering.peak_processing.select_by_intensity module
- matchms.filtering.peak_processing.select_by_mz module
- matchms.filtering.peak_processing.select_by_relative_intensity module
- Submodules
Submodules¶
- matchms.filtering.SpeciesString module
SpeciesStringSpeciesString.dirtySpeciesString.targetSpeciesString.cleanedSpeciesString.__init__()SpeciesString.clean()SpeciesString.clean_as_inchi()SpeciesString.clean_as_inchikey()SpeciesString.clean_as_smiles()SpeciesString.guess_target()SpeciesString.looks_like_a_smiles()SpeciesString.looks_like_an_inchi()SpeciesString.looks_like_an_inchikey()
- matchms.filtering.SpectrumProcessor module
- matchms.filtering.default_filters module
- matchms.filtering.default_pipelines module
- matchms.filtering.filter_order module
